SE (3) diffusion model with application to protein backbone generation
Reciept of FrameDiff
- Train two flow diffusion model: Riemannian diffusion for residue rotation matrices and Gaussian diffusion for residue translations.
- Use best practices from AlphaFold2.
Methodology
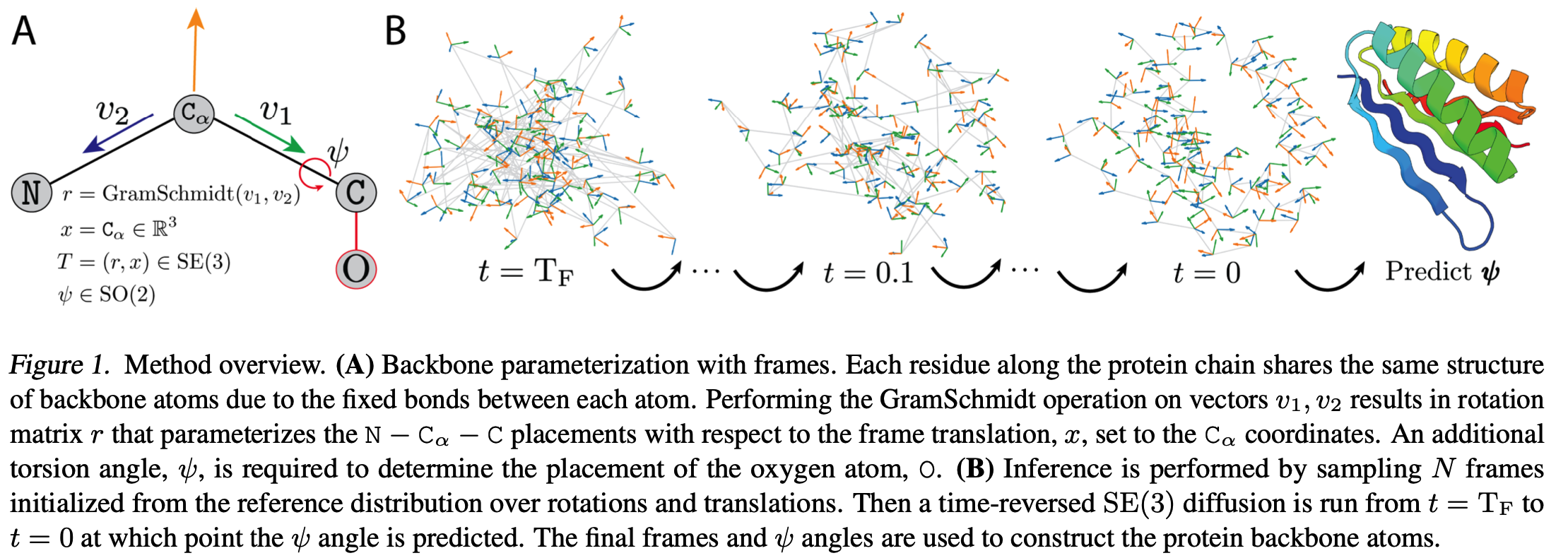
The authors use the same parameterization as in AlphaFold2 and RFDiffusion. The protein is represented as a sequence of residue rotation and translation.
The diffusion process is essentially the same as in RFDiffusion. The main difference is the absence of folding pretraining.
I recommend the presentation of Jason Yim, which compares FrameDiff, RFDiffusion, and FrameFlow.